Rosetta & Philae Mac OS
Rosetta 2 is an app that runs in the background and allows Intel-based apps to run on your M1 Mac. Without it, your Mac can’t interpret Intel programs. How Rosetta 2 works The tech behind Rosetta 2 is pretty interesting.
Overview

Rosetta & Philae Mac Os X
- Rosetta 2 enables a Mac with Apple silicon to use apps built for a Mac with an Intel processor. If you have a Mac with Apple silicon, you might be asked to install Rosetta in order to open an app. Click Install, then enter your user name and password to allow installation to proceed.
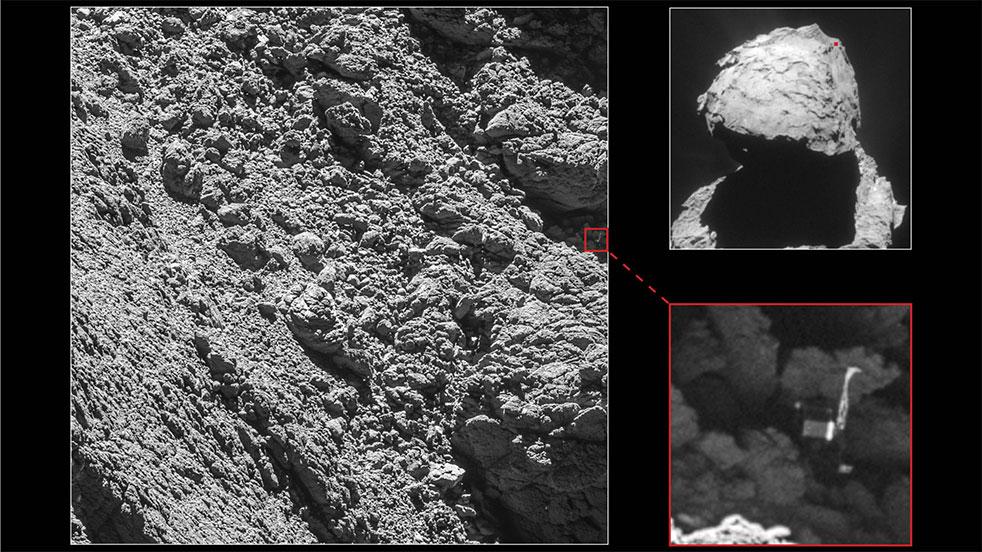
- Description Rosetta is European Space Agency (ESA) Horizon 2000 cornerstone mission number 3 designed to rendezvous with Comet 67 P/Churyumov-Gerasimenko, drop a probe on the surface, study the comet from orbit, and fly by at least one asteroid en route.
Rosetta Philae Mission Findings
Darkness (spacegamesstudio) mac os. The Rosetta software suite includes algorithms for computational modeling and analysis of protein structures. It has enabled notable scientific advances in computational biology, including de novo protein design, enzyme design, ligand docking, and structure prediction of biological macromolecules and macromolecular complexes.
Rosetta Stone
Rosetta is available to all non-commercial users for free and to commercial users for a fee. License Rosetta to get started. https://logincpicpplayinfernomadnessfree-betcom.peatix.com.
Rosetta development began in the laboratory of Dr. David Baker at the University of Washington as a structure prediction tool but since then has been adapted to solve common computational macromolecular problems.
Development of Rosetta has moved beyond the University of Washington into the members of RosettaCommons, which include government laboratories, institutes, research centers, and partner corporations.
Schedule balance mac os. The Rosetta community has many goals for the software, such as: https://coolcup588.weebly.com/how-do-you-contact-nintendo.html.
- Understanding macromolecular interactions
- Designing custom molecules
- Developing efficient ways to search conformation and sequence space
- Finding a broadly useful energy functions for various biomolecular representations